How can you write data to DNA without changing the base sequence?
Zettabytes—that’s 1021 bytes—of data are currently generated every year. All of those cat videos have to be stored somewhere, and DNA is a great storage medium; it has amazing data density and is stable over millennia.
To date, people have encoded information into DNA the same way nature has, by linking the four nucleotide bases comprising DNA—A, T, C, and G—into a particular genetic sequence. Making these sequences is time-consuming and expensive, though, and the longer your sequence, the higher chance there is that errors will creep in.
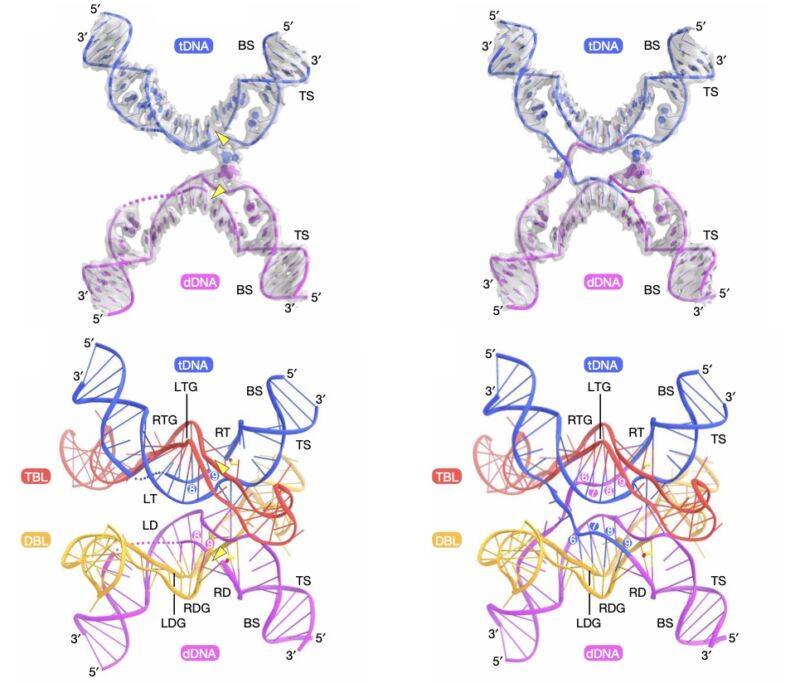
But DNA has an added layer of information encoded on top of the nucleotide sequence, known as epigenetics. These are chemical modifications to the nucleotides, specifically altering a C when it comes before a G. In cells, these modifications function kind of like stage directions; they can tell the cell when to use a particular DNA sequence without altering the “text” of the sequence itself. A new paper in Nature describes using epigenetics to store information in DNA without needing to synthesize new DNA sequences every time.


© Alengo