Study: DNA corroborates “Well-man” tale from Norse saga
A 12th-century Norse saga tells of an invading army from the south razing a castle stronghold and throwing a dead body into the well to render the water undrinkable. Human remains believed to be those of this so-called "Well-man" were discovered in the 1930s, providing valuable potential outside confirmation of the tale. Scientists have now sequenced the DNA of those remains, and while they could not prove once and for all that the remains are those of the Well-man, their findings are consistent with that identification, according to a new paper published in the journal iScience.
Much of what we know about early Norse and Icelandic history comes from the sagas, many of which were written by scholars centuries after the events described—most likely based on oral traditions or earlier now-lost manuscripts. One notable exception is the Sverris Saga, which covers the reign of King Sverre Sigurdsson (1151–1240 CE), a tumultuous period marked by warring factions all vying to claim the throne. Norse scholars think that at least part of this saga was written contemporaneously at the king's request, and it contains detailed descriptions of many battles and speeches and a large cast of characters.
King Sverre's claim to the throne was that he was the son of King Sigurd Munn, killed in 1155 CE by his brother. Sverre's men were known as "Birkenbeiner" because their legwear and shoes were made of birch bark. Among the rival factions were the "Bagleres" from southern Norway. In 1197, King Sverre was spending the winter in Bergen in his stronghold, Sverresborg Castle. Bagler fighters snuck into the castle via a secret door and plundered the place, burning all the homes within the castle walls. That's when they threw a dead man down the local drinking well, subsequently filling the well with boulders.
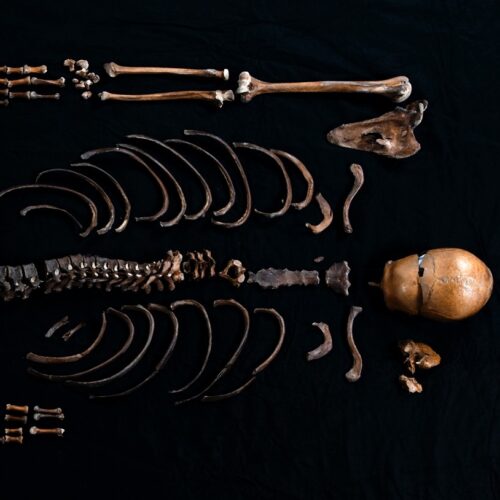
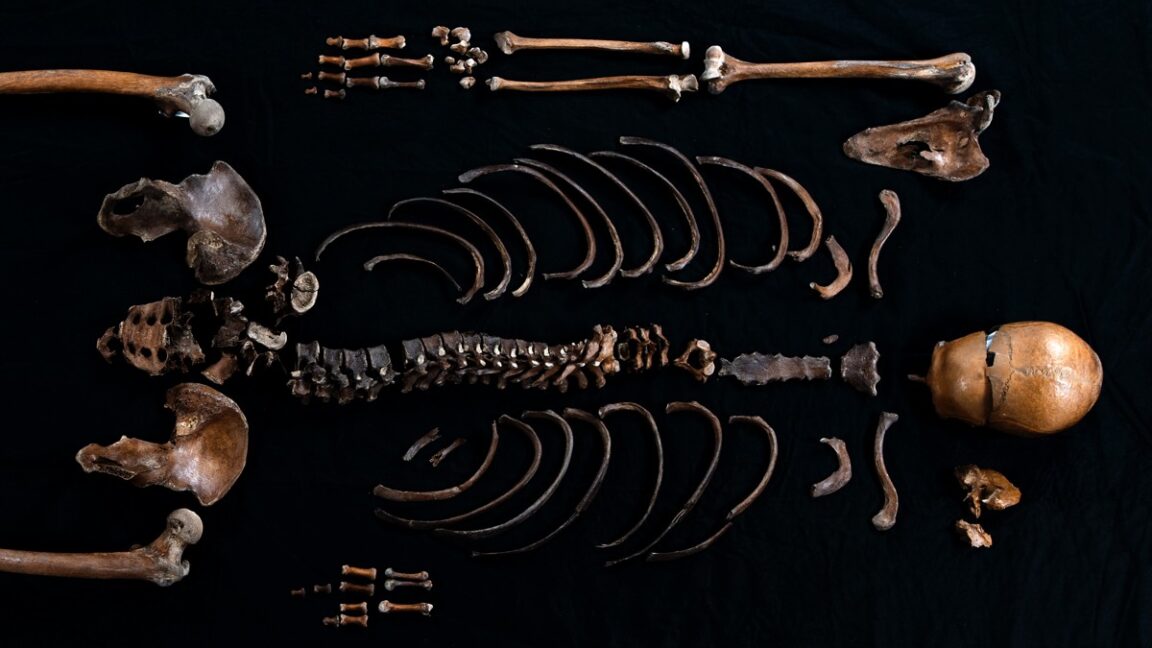
© Åge Hojem NTNU Vitenskapsmuseet/CC BY-SA





